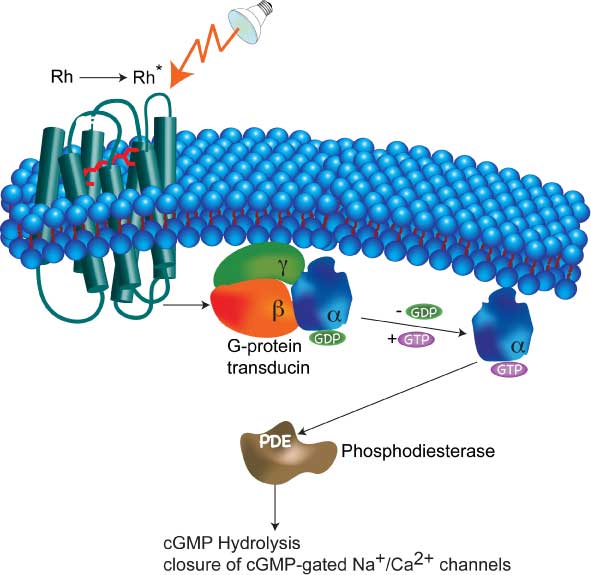
| In addition to conformational fluctuations of proteins and other biomolecules, other dynamically heterogeneous processes occur in the cell. The vast complexity of the cellular interaction networks necessitates unconventional approaches in modeling such processes as signal transduction. Often, only a few molecules are involved in triggering a signaling process, thus, a stochastic description of signal transduction cascades is more appropriate than the traditional macroscopic rate law modeling. In addition, the cellular milieu modifies the dynamics of in vivo binding, thus introducing additional complexity into the stochastic dynamics of signal transduction processes. Using tools from diverse areas of physics and bioinformatics we are building a nonequilibrium statistical mechanics of signal transduction networks. |  |