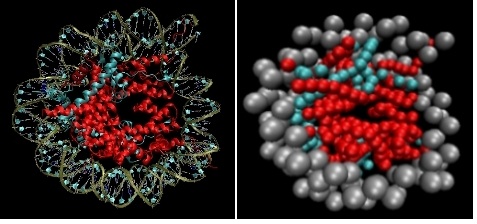
Background. The total length of DNA in each eukaryotic cell can reach 2 m, however, it must be housed in a micrometer size cell. This staggering six orders of magnitude compaction is achieved by wrapping DNA around protein octamers called histones, to form nucleosomes. The latter in turn fold into a superstructure called the 30-nm fiber, which then folds into higher order chromatin structures. Histon octamers consist of a core and flexible protruding tails. Modifications of these tails is thought to regulate the accessability of DNA to transcription factors and the gene transcription machinary.
Theory. A single nucleosome is already an extremely complex system from the molecular point of view: it consists of ~1200 protein residues and ~160 base pairs. Since the nucleosome consists of tens of thousands of atoms, it can not be simulated efficiently with all-atom forcefields. Thus, to model the dynamics of a nucleosomal array or the 30-nm fiber formation we do not explicitly account for all atoms, but group many atoms together into beads in a procedure called “coarse-graining”, and build effective interaction potentials among these beads. Close collaborations with experimental groups helps us to improve our models as well as to test their predictive power. In particular, we are conducting joint theoretical and experimental studies to determine how the nucleosomal structure and dynamics are changed when the histon tails are modified in specific positions.
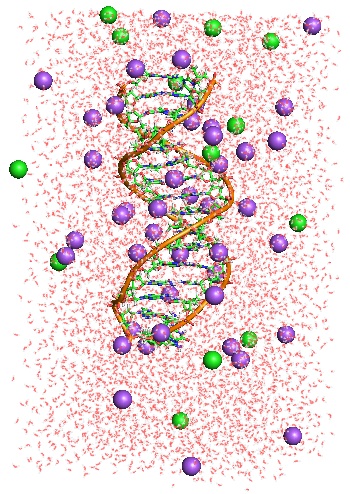
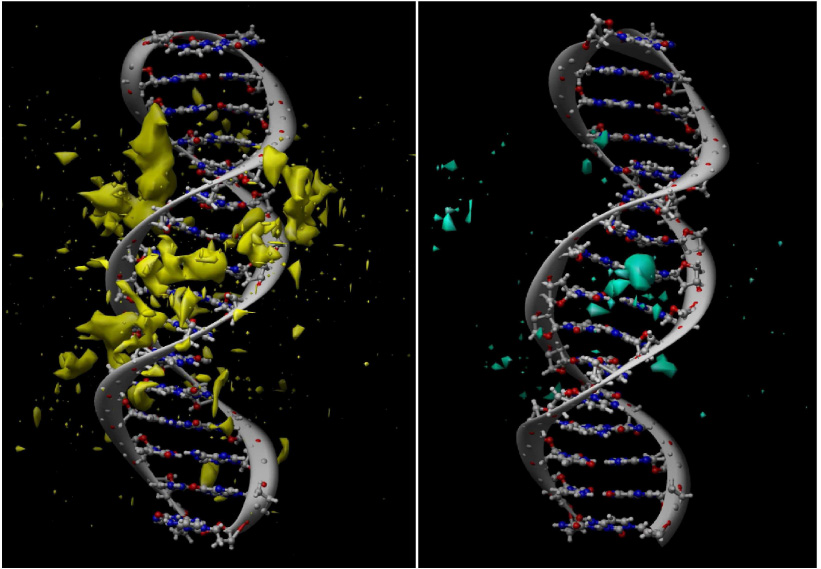
To model poly-nucleosomal arrays we have to achieve better understanding of how DNA interacts with its ionic atmosphere. We have carried out a series of extensive all-atom MD simulations of a 16-base-pair DNA oligomer in different ionic environments. These simulations will be used to develop a coarse-grained model of the linker DNA chain connecting adjacent nucleosomes in a poly-nucleosomal array.